STA workflow
Run the structural tensor analysis (STA) workflow for fiber quantification and tracking.
Attention
Run workflow after running the CLARITY-Allen registration first
Workflow for STA:
Converts Tiff stack to nii incl. down-sampling
Uses registered labels to create a seed mask at the depth (ontology level) of the desired label (seed)
Creates a brain mask
Runs STA analysis using the seed and brain masks
Computes virus intensities for all labels at that depth
Executes:
conv/miracl_conv_convertTIFFtoNII.py
lbls/miracl_lbls_get_graph_info.py
lbls/miracl_lbls_generate_parents_at_depth.py
utils/miracl_extract_lbl.py
utils/miracl_create_brainmask.py
sta/miracl_sta_track_primary_eigen.py
lbls/miracl_lbls_stats.py
Main Outputs
File |
Description |
|---|---|
|
Tract file |
|
Virus stats csv |
|
Streamline density stats csv |
GUI
From the main GUI (invoked with: $ miraclGUI), select Workflows ->
CLARITY STA:
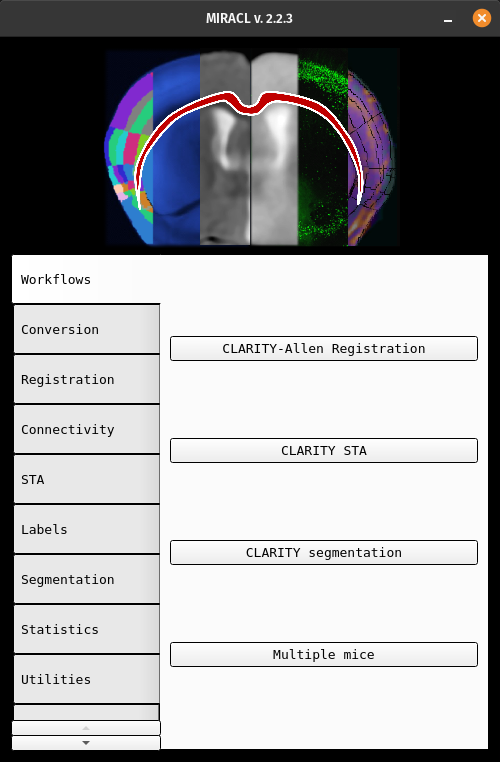
The following window will appear:

Hint
To open the STA workflow menu directly use: $ miracl flow sta
Click on Select Input tiff folder and choose the folder that contains the
virus channel from the dialog window.
Then choose the registered Allen labels inside the final registration folder
(reg_final) from the dialog window by clicking on Select CLARITY final
registration folder.
Next choose the output file name (Output nii name), e.g. Mouse05. Our
script will automatically append downsample ratio and channel info to the given
name.
Set the tracking parameters:
Parameter |
Description |
Default |
|---|---|---|
Seed label abbreviation |
From Allen atlas ontology, for the seed region. Examples: Combined hemispheres:
Right hemisphere:
|
Required. Function will exit with error 1 if not provided. |
hemi |
Labels hemisphere. Accepted inputs are:
|
|
Derivative of Gaussian (dog) sigma |
Example: |
|
Gaussian smoothing sigma |
Example: |
|
Tracking angle threshold |
Example: |
|
Use 2nd order runge-kutta method for tracking |
Use 2nd order runge-kutta:
|
|
And the tiff conversion parameters:
Parameter |
Description |
Default |
|---|---|---|
Downsample ratio |
Set the downsample ratio. |
|
chan # |
For extracting single channel from multiple channel data. |
|
chan prefix |
String before channel number in file name. Example: |
|
Resolution (x,y) |
Original resolution in x-y plane in um. |
|
Thickness |
Original thickness (z-axis resolution/spacing between slices) in um. |
|
Downsample in z |
Downsample in z dimension. Binary:
|
|
Users can also input their own brain mask, as well as their own seeding mask.
Both masks would respectively replace the automatically generated brain mask
and regional mask used for the tractography. Users also have the option to
dilate the seed mask across any of the three dimensions, by a value (indicated
by the Dilation factor fields).
Attention
Note that the following parameters are required:
tiff folderoutput nii nameSeed label abbreviationCLARITY final registration folderhemiDerivative of GaussianGaussian smoothing sigmaTracking angle threshold
After choosing the parameters, first press Enter to save them and then
Run to start the workflow.
Command-line
Usage:
$ miracl flow sta -f [ Tiff folder ] -o [ output nifti ] -l [ Allen seed label ] -m [ hemisphere ] -r [Reg final dir] -d [ downsample ratio ]
Example:
$ miracl flow sta -f my_tifs -o clarity_virus -l PL -m combined -r clar_reg_final -d 5 -c AAV g 0.5 -k 0.5 -a 25
Or for right PL:
$ miracl flow sta -f my_tifs -o clarity_virus -l RPL -m split -r clar_reg_final -d 5 -c AAV -g 0.5 -k 0.5 -a 25
Arguments:
arguments (required):
-f: Input Clarity tif folder/dir (folder name without spaces)
-o: Output nifti
-l: Seed label abbreviation (from Allen atlas ontology)
-r: CLARITY final registration folder
-m: Labels hemi
-g: Derivative of Gaussian (dog) sigma
-k: Gaussian smoothing sigma
-a: Tracking angle threshold
optional arguments:
-d: Downsample ratio (default: 5)
-c: Output channel name
-n: Chan number for extracting single channel from multiple channel data (default: 0)
-p: Chan prefix (string before channel number in file name). ex: C00
-x: Original resolution in x-y plane in um (default: 5)
-z: Original thickness (z-axis resolution/spacing between slices) in um (default: 5)
-b: Brain mask (to replace brain mask automatically generated by workflow)
-u: Seed mask (in place of regional seed mask generated by workflow)
-s: Step length
--downz: Downsample in z
--dilationfx: Dilation factor for x (factor to dilate seed label by)
--dilationfy: Dilation factor for y (factor to dilate seed label by)
--dilationfz: Dilation factor for z (factor to dilate seed label by)
--rk: Use 2nd order range-kutta method for tracking (default: 0)
--out_dir: Output directory
Jupyter notebook
An accompanying Jupyter notebook for this tutorial can be found here.